Accelerate your research with superior assay technologies and overcome the challenge of selecting the "right" target antigen!
T Cell Epitope Mapping
Significantly improve your peptide epitope discovery process, slash development costs and strengthen your intellectual property portfolio by accessing our most advanced combinatorial peptide library screening approaches for vaccine, therapeutic and diagnostic developments.
Introduction to T Cell Epitope Mapping
T cell epitope mapping has emerged as one of the most powerful new drug discovery tools for a range of biomedical applications. With this growing interest, Pure Protein has developed a comprehensive and novel discovery program aimed at identifying and selecting peptides, capable of generating adequate immune mobilization for vaccine, therapeutic and diagnostic modalities.
The wealth of newly available genomic sequence information provides a superb source for the identification of novel epitope targets deriving from tumor-specific proteins, from proteins of viral, bacterial or other pathogenic origins, as well as from host proteins that are uniquely expressed, processed, modified or degraded during infections or cancerous states. Our services will help in decreasing the impact of diseases for which effective vaccines or therapies are lacking or for which existing therapies are incompletely effective and/or even toxic to humans.
How it Works
Select target protein, library modality and HLA alleles
Select a specific target from cancer antigens or pathogenic sequence that provides the source for your novel epitopes and choose a library modality to be specifically synthesized and screened with the HLA allele combination of your choice to identify the most potent T cell epitope candidates for your discovery program.
Receive high-throughput screening hit report
Receive primary screening results identifying candidates with affinities ranging from high to low using our state-of-the-art peptide screening assay. Moreover, the assay eliminates all non-binding peptides which need not be investigated further. The use of our technology in this primary screening will speed up your lead discovery process and deliver higher quality hits.
Prioritize top candidates for peptide epitope validation
High affinity binding is the critical factor controlling immunogenicity of peptides. Within this refinement process, prioritize the highest ranked screening hits and we validate their accurate potential by generating dose–response curves determining their inhibitory concentration (IC50) as measure of their effectiveness.
Receive a mapping report
After completion of the screens, the first epitope map for a cancer or virus-related protein can be created, showing detailed information on position of each epitope on the sequence string, their HLA-type and screening score. In addition, the map uncovers the location of immunological hotspots and cross-reactivity patterns among all HLA-molecules investigated.
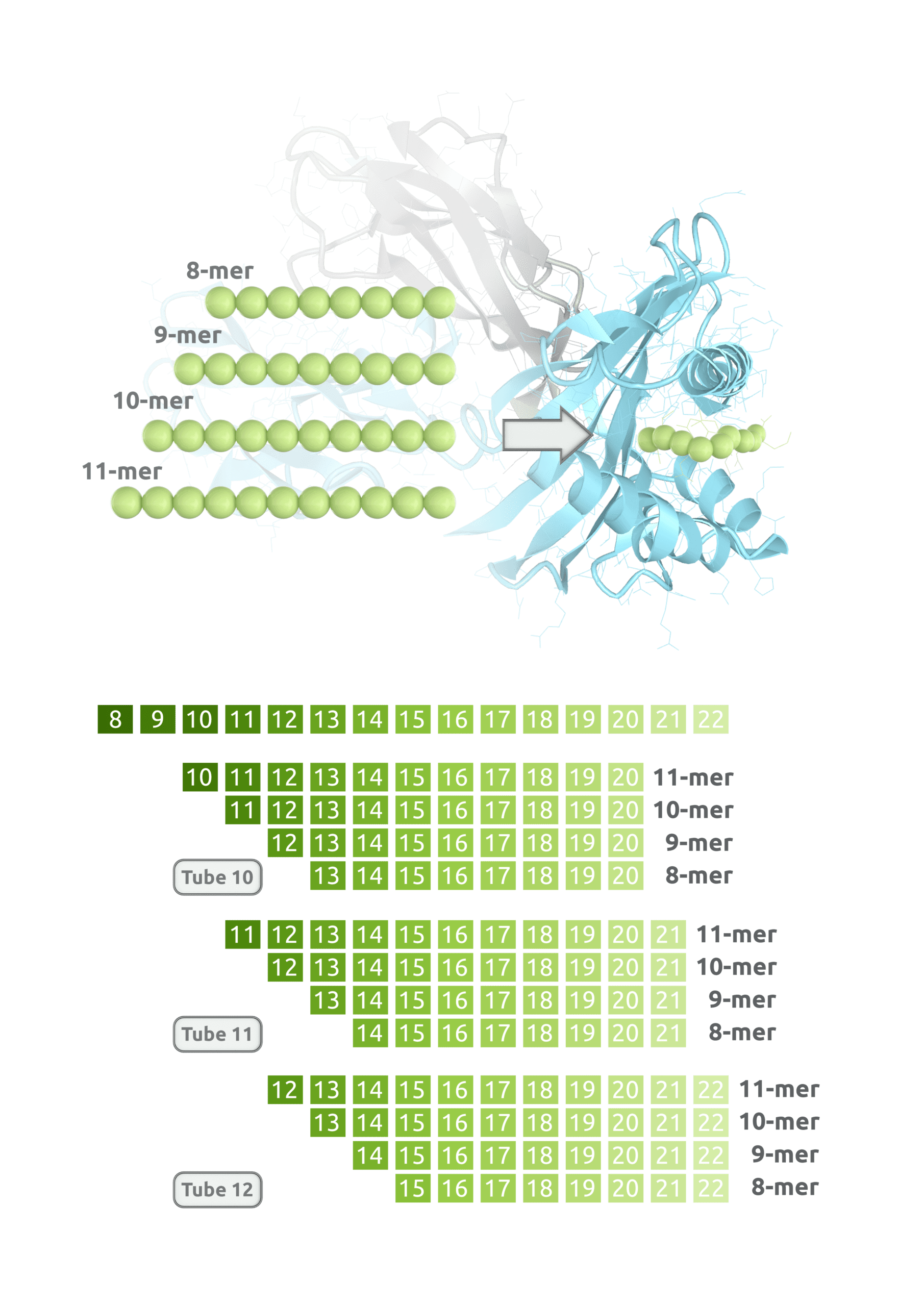
Mapping Concept
Never miss an epitope! A truncation library of overlapping peptides with systematic truncation of the flanking residues allows the screening of any possible combination of 8, 9, 10, and 11-mer of a chosen target protein.
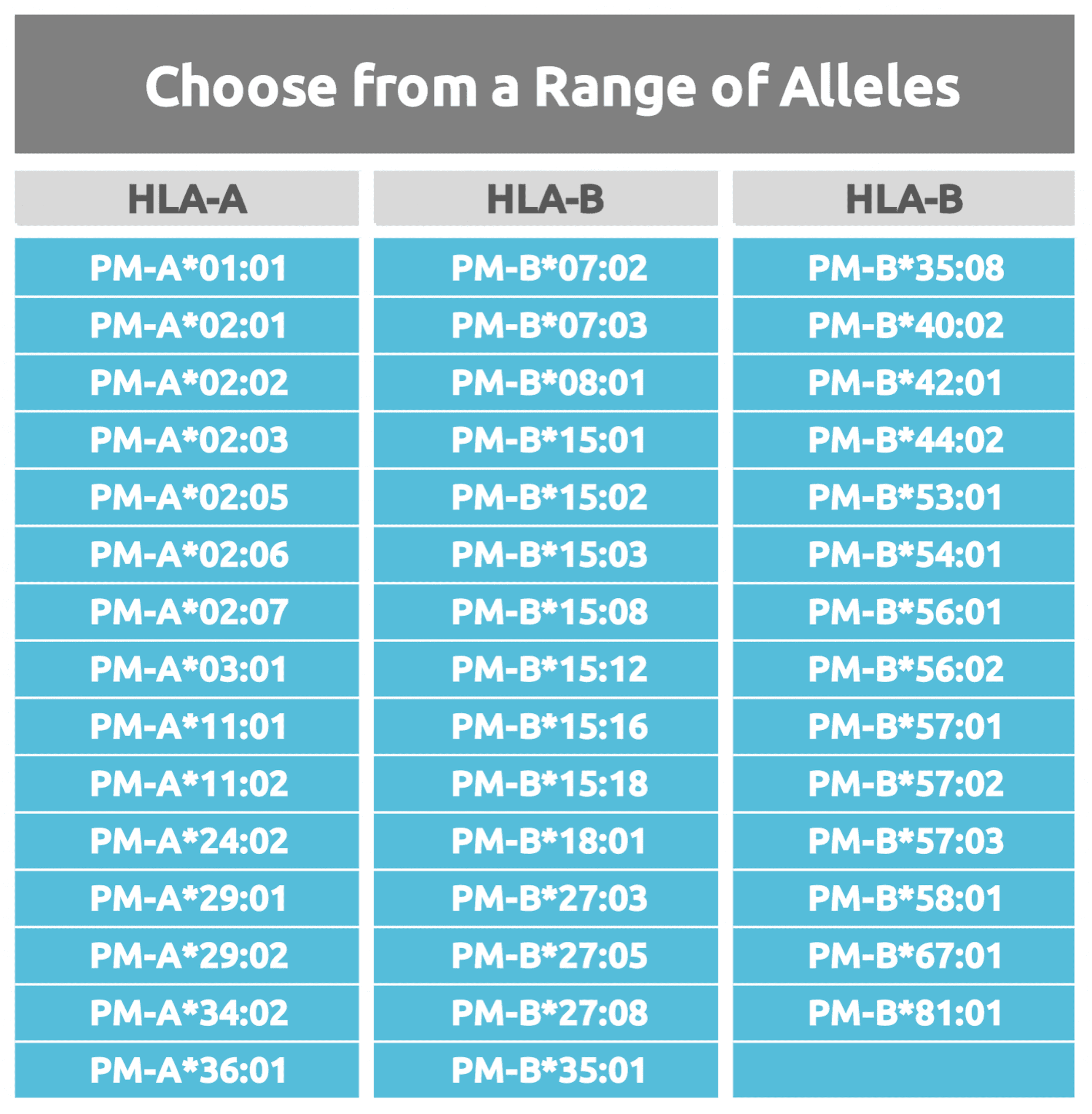
T cell Epitope Mapping Service
Download our list of HLA alleles available for our T cell Epitope Mapping Service. View all available proteins as well as services in development.
Target Selection
Knowledge of proteins that are being targeted by the immune system is very valuable information for the development of vaccines, therapeutic interventions and diagnostics.
CANCER TARGET SELECTION
Select from the wealth of newly available genomic sequence information that provides a superb source for the identification of novel peptide epitope targets deriving from cancer antigens or other aberrant gene expressions, or from host proteins that are uniquely expressed, processed, modified during cancerous states.
INFECTIOUS DISEASE TARGET SELECTION
Select from the wealth of pathogenic sequences that provides the source for the identification of novel peptide epitope targets deriving from viral, bacterial or other pathogenic origins, or from host proteins that are uniquely expressed, processed, modified or degraded during infections.
Library Selection
In the discovery of new T cell epitopes, libraries of overlapping peptides can be used to identify the binding capability to a broad range of HLA alleles to select the most potent T cell epitope candidates.
TRUNCATED LIBRARY SELECTION
“Never miss an epitope!” Select a truncation library of overlapping peptides with systematic truncation of the flanking residues allowing the screening of any possible combination of 8, 9, 10, and 11-mer of a chosen target protein for a seamless HLA Class I epitope discovery assuring that no epitope is left behind to produce effective immune responses.
OVERLAP LIBRARY SELECTION
Prioritize your discovery! Our overlapping peptide library approach is used for linear, continuous epitope mapping, with the aim to generate an overlapping peptide sequence of specific length and single offset to cover the entire native protein sequence. Select the most popular libraries using 9mers or any other length of choice for your HLA Class I discovery.
Deliverables
The primary goal of epitope discovery has been to identify T cell peptide epitopes for use in the construction of vaccines that activate a clinically relevant cellular immune response to treat or prevent various types of infectious diseases and cancers. The peptide epitope mapping system is a high-throughput assessment tool that characterizes peptides derived from various source libraries and offers the opportunity to study thousands of potential epitopes in a single experiment across multiple alleles. This highly advanced system not only identifies screening hits but properly prioritizes these potential T-cell epitopes according to their HLA affinity.
The Epitope Map
To effectively select for high, medium and low affinity binders among the HLA types selected, a single reaction contains a set of four truncated peptides in sizes between 8-11 amino acids. With our truncated, overlapping peptide mixes, we are able to provide a comprehensive picture of epitopes for a seamless HLA Class I epitope discovery assuring that no epitope is left behind to produce effective immune responses. These maps show detailed information on the position of each epitope on the sequence string, their HLA-type, and screening score. Identification of positive hits within the pool of four will be resolved in a follow-up screen.
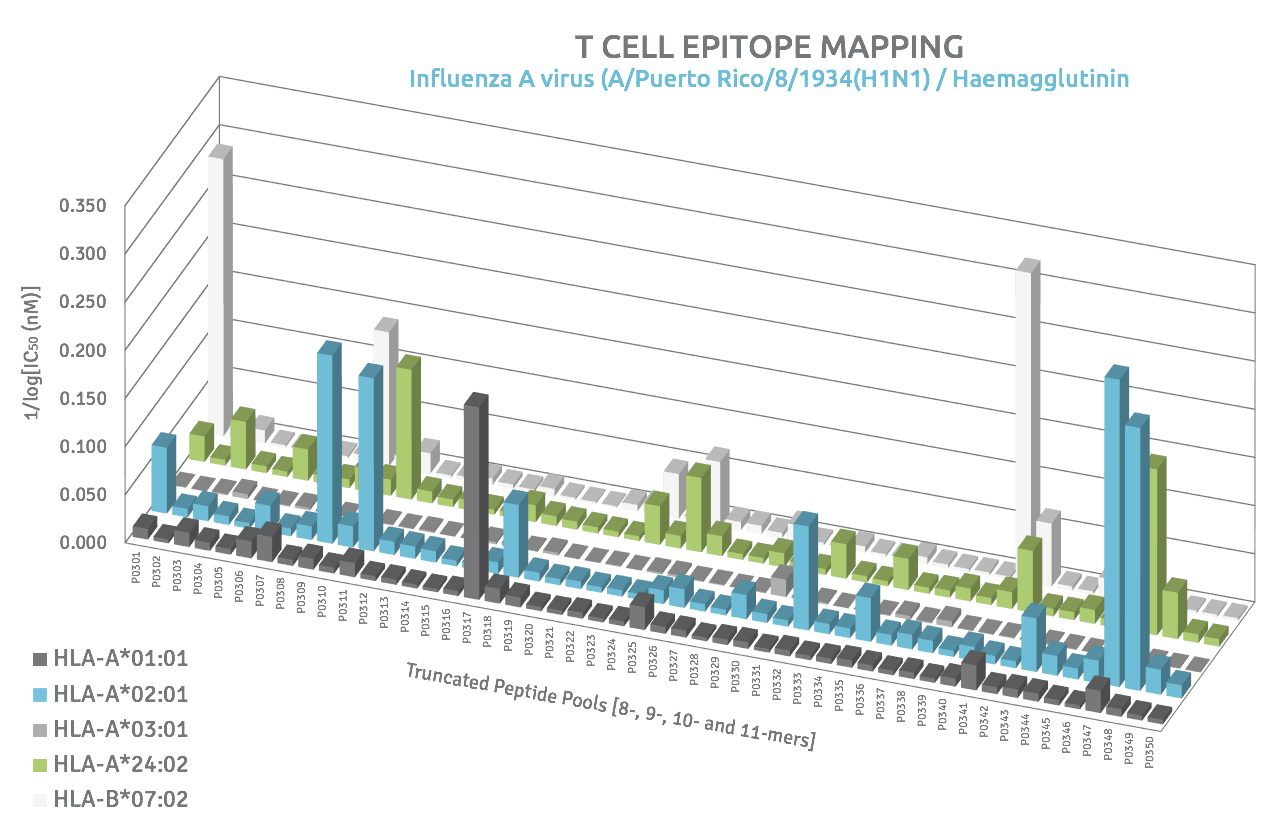
Cross-Reactivity Patterns
Cross-reactivity is explained as the close resemblance of subsets of HLA proteins in the structure of the peptide-binding groove. Classification of allele overlapping peptide binding specificities has become an important issue in vaccine design with direct implications concerning population coverage. Our mapping concept easily identifies such cross-reactive peptides and allows to draw conclusions about their usefulness for treating a larger subset of patients.
Immunogenic Hotspot
T cell epitope mapping approach is a great tool in the identification of immunogenic hotspots which is based on recent findings that suggest that HLA ligands are not randomly distributed along the proteins’ sequences but are located within “hotspots”, which fit proteasomal cleavage, peptide processing and HLA-binding rules. These immunological hotspots may be more stable polypeptides that better survive the highly proteolytic cytosolic environment of living cells and therefore provide focus points for T cell epitope discovery in the context of cross presentation of multiple alleles. Immunogenic hotspots will provide critical additional information to drive epitope prioritization which will have a substantial impact on the selection of T cell peptide epitopes for vaccination.
Frequently Asked Questions
What do I get when I order a Mapping Service?
Epitope maps for a cancer or virus-related protein will be created, showing detailed information on position of each epitope on the sequence string, their HLA-type and screening score. In addition, the map uncovers the location of immunological hotspots and cross-reactivity patterns among all HLA-molecules investigated.
How do I start a mapping project?
If you have questions or are looking for some help in defining your project needs, reach out to our experts by visiting the Contact Us page or write us at [email protected]. One of our team members will be sure to get back to you in no time.
What is a Truncation Library?
A Truncation Library consists of overlapping peptides with systematic truncation of the flanking residues allowing the screening of any possible combination of 8, 9, 10, and 11-mer of a chosen target protein for a seamless HLA Class I epitope discovery assuring that no epitope is left behind to produce effective immune responses.
Talk to an expert today
Want to know more about our Mapping Systems?
If you’re still wondering if our services are the right technology to take you to the next level and are looking for some help in defining your project needs and customize to the most relevant assay solution, reach out to our experts by visiting the Contact Us page. Someone from our team will get back to you in no time.
Expert advice, whenever you need it!
Related Products & Services
Soluble HLA-B
Select from a pool of over 65 HLA-B alleles to drive your immunological research to identify antibody immune responses or visualize antigen-specific immune cells.
Soluble HLA-C
Discover a selection of over 20 HLA-C alleles, the dominant ligand for KIR on NK cells for your immunological research.
Peptide Epitope Validation
Validate individual peptide epitope candidates by determining their physical binding characteristics and directly measure accurate IC50 values in order to judge and properly prioritize their immunogenic potential.

